Files involved in RNA-seq
Here, I learnt about all the different files that may come up during a NGS analysis. All the knowledge comes from:
General:
- https://learn.gencore.bio.nyu.edu A very useful website teaching about NGS analysis.
FASTA & FASTQ:
- https://support.illumina.com/bulletins/2016/04/fastq-files-explained.html
- http://www.biotrainee.com/thread-2703-1-2.html
- https://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastDocs&DOC_TYPE=BlastHelp
SAM/BAM/CRAM:
GFF/GTF:
- https://useast.ensembl.org/info/website/upload/gff.html
- https://en.wikipedia.org/wiki/Gene_transfer_format
Unix
FASTQ and FASTA
The first section is about FASTQ and FASTA files.
FASTQ
FASTQ files contains four lines including:
- The sequence Identifier with information about the sequencing run and the cluster.
- The sequence read.
- A separator, which is simply a plus (+) sign, but sometimes might also be a (-) sign.
- The quality scores. Using ASCII characters to represent the numerical quality of the scores.
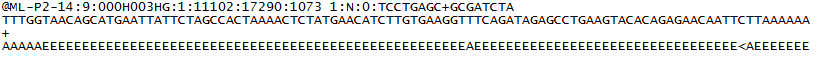
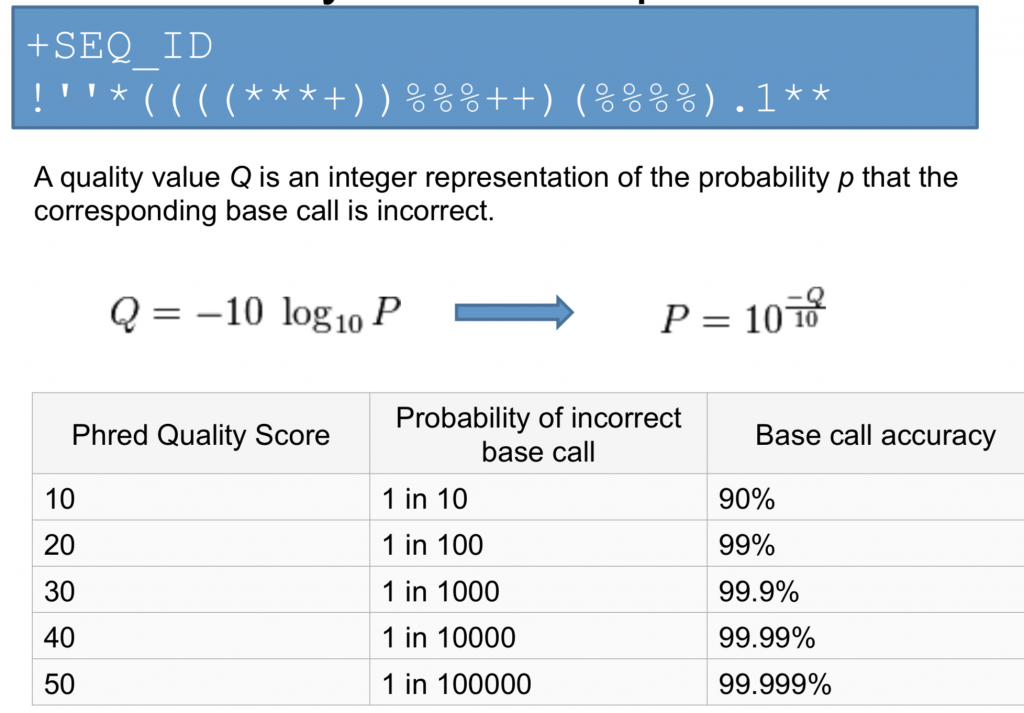
Quality Scores
Quality scores are a way to assign confidence to a particular base within a read. The code is related to the ASCII table and each letter is related to a different Q score.
The quality score Q is related to the probability of incorrect corresponding base call.
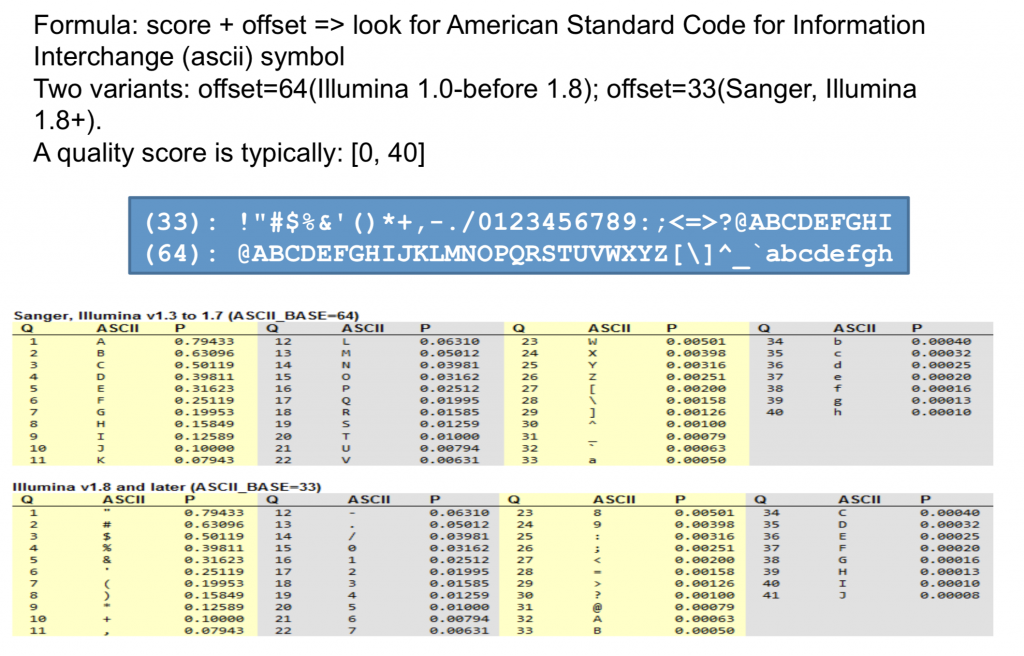
By matching the ASCII table characters to their Q code under certain protocals, we can easily (?) know the quality of the specific gene.
FASTA
FASTA format is a basic format for repoting a sequence whether it's a nucleic acid or amino acid. Made up by two parts:
- A sequence header which starts with a '>' and contaisn the sequence description.
- The sequence itself (containing no space within).
>P01013 GENE X PROTEIN (OVALBUMIN-RELATED) QIKDLLVSSSTDLDTTLVLVNAIYFKGMWKTAFNAEDTREMPFHVTKQESKPVQMMCMNNSFNVATLPAE KMKILELPFASGDLSMLVLLPDEVSDLERIEKTINFEKLTEWTNPNTMEKRRVKVYLPQMKIEEKYNLTS VLMALGMTDLFIPSANLTGISSAESLKISQAVHGAFMELSEDGIEMAGSTGVIEDIKHSPESEQFRADHP FLFLIKHNPTNTIVYFGRYWSP
SAM/BAM/CRAM Format
SAM
The Sequence Alignment/Map (SAM) format is the most basic and most human readable format of the three. It consists of a header, and a row for every read in your dataset, and 11 tab-delimited fields describing that read.
SAM Header
The SAM header varies in size, but header lines start with '@', while alignment lines do not. You can customize the header when you generate the SAM file, add the information you decided to add. The full list of header fields are found below. '*' means that this tag is required; e.g., every @SQ header line must contain SN and LN fields.
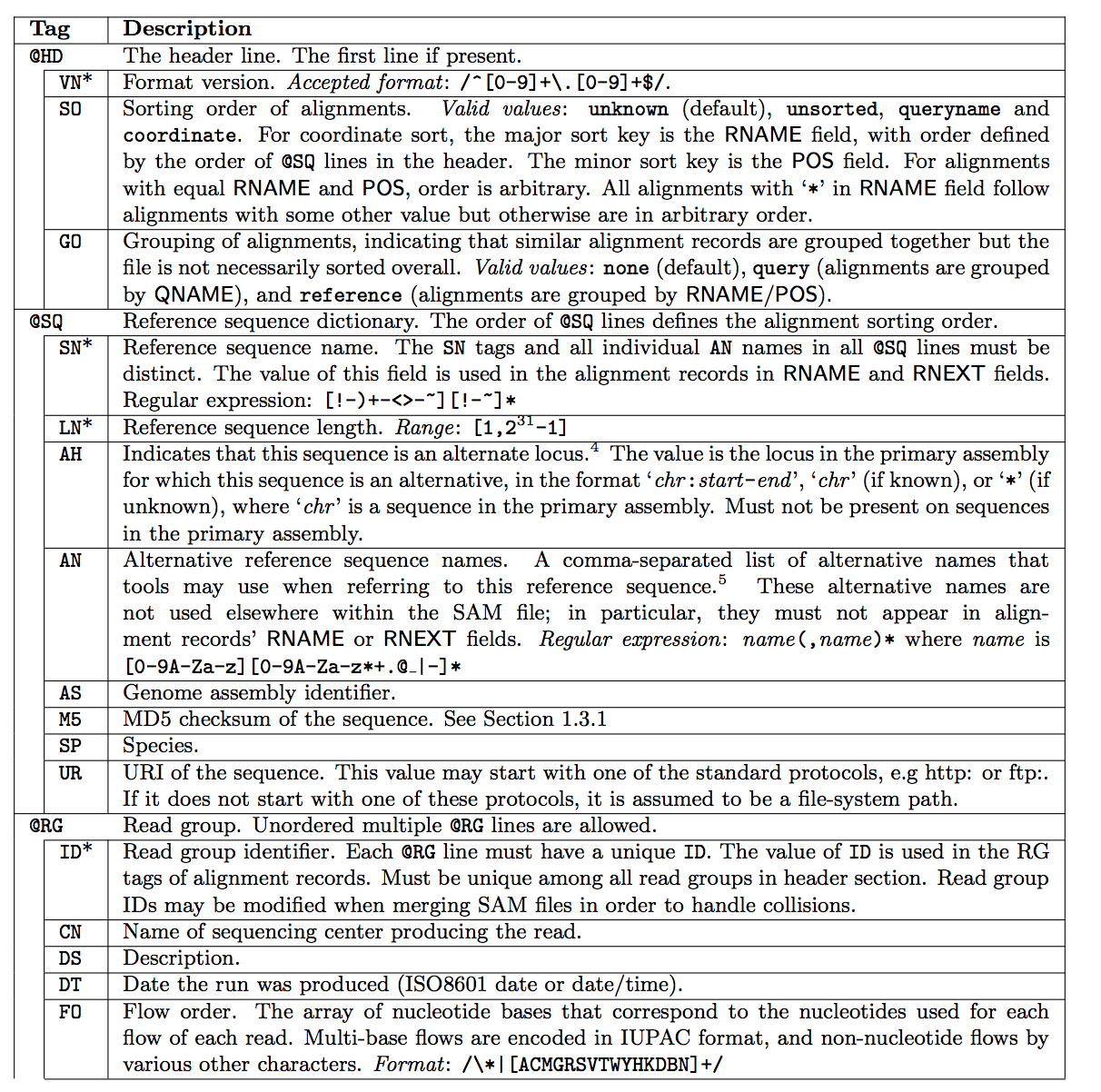
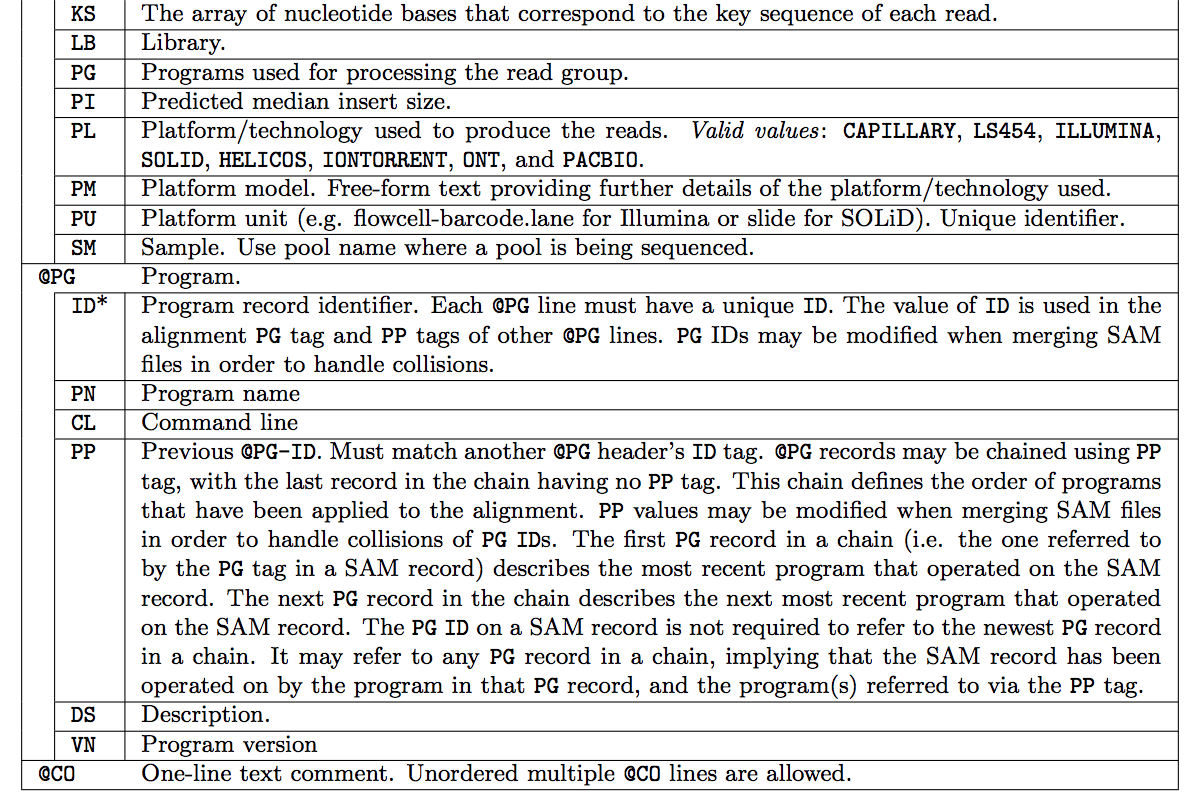
Fields Descriptions
Every row contains 11 mandatory fields, which are:
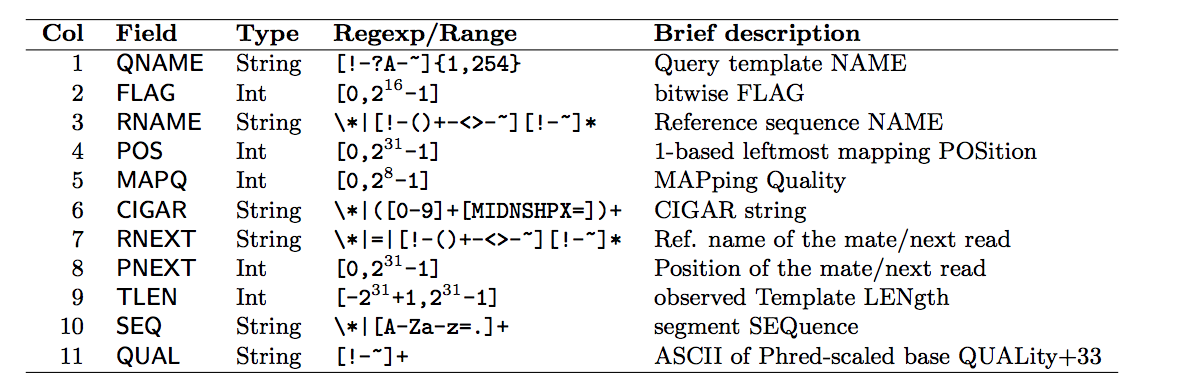
Bitwise Flag is a lookup code to explain certain features about the particular read (exact same concept as Linux permission codes!). It tells you whether the read aligned, is marked a PCR duplicate, if it’s mate aligned, etc. and any combination of the available tags, seen below:
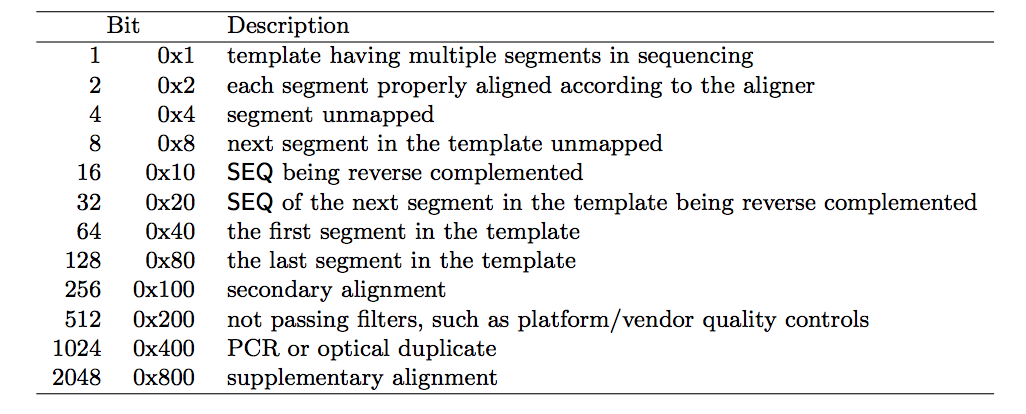
Sometimes it might be a bit hard to explain the integers of the Bitwise Flag, you can go on Picard to get a quick analysis.
Map Q (Mapping Quality) reveals how well the read are aligned to the reference. Different algorithm might lead to different scores, but generally, the greater the number, the better the alignment.
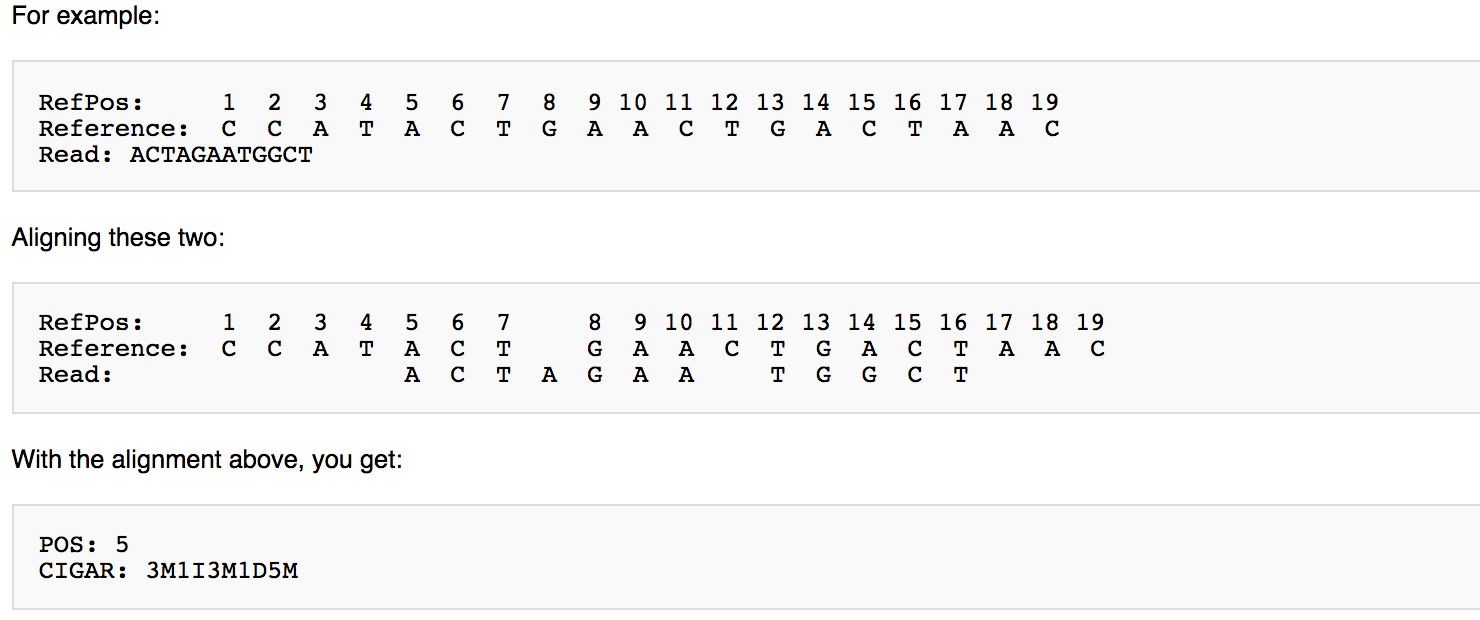
CIGAR String is a special string that can tell you the alignment information of the whole sequence.

BAM
BAM format has literally the same content as the SAM file, except it's in Binary format. Thus it's not legible to human but is smaller and faster to read for a computer.
Tools like Samtools, Picard Tools and IGV are required to make sence of BAM files.
CRAM
This is a relatively new format that is very similar to BAM as it also retains the same information as SAM and is compressed, but it is much smarter in the way that it stores the information. It’s very interesting and up and coming but is a bit beyond my level and not so relative to RNA-seq. To learn more about it, you can read this.
VCF Format
Variant calling format is a tab-delimited text file (different from Virtual Card Format), it is used to describe single nucleotide variants (SNVs) as well as insertions, deletions and other sequence variations. It is limited to only show the variations not the genetic features.
- Chromosome Name
- Chromosome Position
-
ID
This is generally used to reference an annotated variant in dbSNP or other curate variant database.
-
Reference base(s)
- Alternate base(s)
-
Variant quality
Phred-scaled quality for the alternate base. Usually, a >20 score is acceptable.
-
Filter
Whether or not this has passed all filters – generally a QC measure in variant calling algorithms
-
Additional Information
This is for additional information, generally describing the nature of the position/variants with respect to other data.
For example vcf files, look at your own files during the class BIOS201, Genome, why are we different.
GFF and GTF
Gene transfer format (GTF) and General feaure format (GFF) are kinds of file format used to hold information about gene structure.
https://github.com/The-Sequence-Ontology/Specifications/blob/master/gff3.md
BED
The formal bed format description can be found here.
In shorter words, BED files allows an easy way to define the things you want to show in your track annotation (like a reference genome).
Bed Graph
The detailed discription for bed graph is here, I think this file can be used to store the reads for each small sets of nucleotide on your chromosome after your sequencing.
BigWig
The BigWig files indicates the reads on a certain position. You can view it with igv, but also other applications and can turn it into a very beautiful graph. Requires further inquiry. The official BigWig file explaination can be found here.